Estimating the potential energy surface of molecular Hydrogen with ZNE and PennyLane + Cirq#
In this example we apply zero-noise extrapolation (ZNE) to the estimation of the potential energy surface of molecular Hydrogen (\(\rm H_2\)). The variational algorithm used in this example is based on O’Malley et al. PRX (2016) [62].
With the appropriate settings (see code comments in the ZNE subsection), this notebook reproduces the results shown in Figure 4 of the Mitiq white paper.
Figure 1: Pictorial representation of the \(\rm H_2\) molecule. In this example, we variationally estimate the potential energy surface of the molecule as a function of the bond length \(R\).#
from functools import partial
import matplotlib.pyplot as plt
import numpy as np
from scipy.optimize import brute
import cirq
import pennylane as qml
from pennylane.pauli import pauli_word_to_string
import mitiq
from mitiq import zne # Zero-noise extrapolation module
from mitiq.interface.mitiq_cirq import compute_density_matrix
The Hamiltonian for \(\rm H_2\)#
We can write down the Hamiltonian for \(\rm H_2\) in the following form [62]
where \(g_i\) are numerical values that depend on the bond length \(R\). The above Hamiltonian:
Uses the minimal STO-6G basis,
Uses the Bravyi-Kitaev transform, and
Reduces resources (qubit number) by symmetry considerations (see [62] for more details).
Each coefficient \(g_i\) is a function of the bond length \(g_i = g_i(R)\). We obtain the H for \(\rm H_2\) at any given bond length \(R\)
by first building the full molecular Hamiltonian using PennyLane and then
tapering it using the symmetries mentioned in the Appendix A of [62] to reduce it to a two qubit Hamiltonian,
where the qubits 0 and 2 corresponds to the labels 0 and 1 in the Hamiltonian H given above.
ang_to_bohr = 1.8897259886 # pennylane specifies coordinates in terms of Bohr radius
def molecular_hamiltonian(bond_length: float) -> mitiq.Observable:
""" Builds the Hamiltonian for H2 at a given bond length """
symbols = ['H', 'H']
geometry = ang_to_bohr * np.array([[0., 0., - 0.5 * bond_length],
[0., 0., 0.5 * bond_length]])
mol = qml.qchem.Molecule(symbols, geometry)
Hamil, qubits = qml.qchem.molecular_hamiltonian(symbols, geometry, basis="sto-6g",
method="pyscf", mapping="bravyi_kitaev")
# tapering the Hamiltonian using symmetries
generators = [qml.PauliZ(1), qml.PauliZ(3)] # symmetries corresponding to qubit 1 and 3
paulix_ops = [qml.PauliX(1), qml.PauliX(3)] # corresponding paulix ops for tapering
eigval_sec = [1, 1] # optimal sector containing the ground state
Hamil_tapered = qml.taper(Hamil, generators, paulix_ops, eigval_sec)
# converting the tapered Hamiltonian to mitiq Observable
coeffs, ops = Hamil_tapered.terms()
wire_map = {0 : 0, 2 : 1} # for relabeling the qubits in the Pauli representation
pauli_str = [mitiq.PauliString(
pauli_word_to_string(op, wire_map=wire_map), coeff=coeff)
for coeff, op in zip(coeffs, ops)]
hamil_obs = mitiq.Observable(*pauli_str)
return hamil_obs
radii = np.linspace(0.2, 2.6, 13)
hamiltonians = [molecular_hamiltonian(rad) for rad in radii]
print(f"Hamiltonian at R = {radii[0]} Angstroms is:\n {hamiltonians[0]}")
Hamiltonian at R = 0.2 Angstroms is:
(2.848902703661773+0j)*I + (0.5678139466276169+0j)*Z(q(0)) + (-1.4507698908878524+0j)*Z(q(1)) + (0.0790693822994611+0j)*Y(q(0))*Y(q(1)) + (0.0790693822994611+0j)*X(q(0))*X(q(1)) + (0.6799394856539501+0j)*Z(q(0))*Z(q(1))
We define a function that evaluates the expectation value of the Hamiltonian on a noisy backend and for a given input circuit (ansatz). Here we use a Cirq density matrix simulator with a simple depolarizing noise model.
def energy(
ansatz: cirq.Circuit, radius_index: int, depo_noise_strength: float = 0.05
) -> float:
"""Computes the energy at a given bond length (radius)."""
hamiltonian = hamiltonians[radius_index]
executor = mitiq.Executor(compute_density_matrix)
kwargs = {"noise_model_function":cirq.depolarize, "noise_level":(depo_noise_strength,)}
expec_val = executor.evaluate(ansatz, hamiltonian, **kwargs)[0]
return expec_val.real
Evaluating the energy landscape for a fixed \(R\)#
We first define a function that returns single-parameter variational circuit. The ansatz is based on on Fig. 1 of [62].
def ansatz(theta: float) -> cirq.Circuit:
"""Returns the circuit associated to the input variational parameter."""
qreg = cirq.LineQubit.range(2)
return cirq.Circuit(
cirq.ops.X.on(qreg[1]),
cirq.ops.ry(np.pi / 2).on(qreg[0]),
cirq.ops.rx(-np.pi / 2).on(qreg[1]),
cirq.ops.CNOT.on(*qreg),
cirq.ops.rz(theta).on(qreg[1]),
cirq.ops.CNOT.on(*qreg),
cirq.ops.ry(-np.pi / 2).on(qreg[0]),
cirq.ops.rx(np.pi / 2).on(qreg[1]),
)
Below we set a particular radius \(R\) and sweep the \(\theta\) parameter to evaluate the unmitigated energy landscape \(E(\theta, R)\) using different strengths \(p\) of depolarizing noise.
# Noise levels
pvals = (0.00, 0.02, 0.04)
# Variational parameters
thetas = np.linspace(0.0, 2.0 * np.pi, 10)
all_energies = []
for pval in pvals:
energies = []
for theta in thetas:
energies.append(energy(ansatz(theta), radius_index=0, depo_noise_strength=pval))
all_energies.append(energies)
The unmitigated energy landscapes are now stored in the all_energies variable and will be visualized
later.
Applying zero-noise extrapolation to estimate \(E(\theta, R)\)#
We now use zero-noise extrapolation to error-mitigate the energy landscapes \(E(\theta, R)\) for a fixed value of \(R\) and for different levels of the noise.
# Set noise scaling method
scaling_function = zne.scaling.fold_global
# Set extrapolation method
fac = zne.inference.RichardsonFactory(scale_factors=[1, 3, 5])
# Set number of trials per expectation value
num_to_average = 1
# To reproduce the results of the Mitiq paper use the following settings
# scaling_function = zne.scaling.fold_gates_at_random
# pfac = zne.inference.PolyFactory(order=3,
# scale_factors=[1., 1.5, 2., 2.5, 3., 3.5, 4., 4.5, 5., 5.5, 6.])
# num_to_average = 5
# pvals = (0.00, 0.02, 0.04, 0.06)
# thetas = np.linspace(0.0, 2.0 * np.pi, 20)
all_mitigated = []
for p in pvals[1:]:
mitigated = []
for theta in thetas:
# Define an executor function
execute = partial(energy, radius_index=0, depo_noise_strength=p)
# Run ZNE
zne_value = zne.execute_with_zne(
ansatz(theta),
execute,
factory=fac,
scale_noise=scaling_function,
num_to_average=num_to_average,
)
mitigated.append(zne_value)
all_mitigated.append(mitigated)
In the next figure we visualize the following results:
The ideal noiseless energy landscape \(E(\theta, R)\) corresponding to
all_energies[0];The noisy (unmitigated) energies
all_energies;The corresponding error-mitigated energies
all_mitigated.
plt.rcParams.update({"font.family": "serif", "font.size": 14, "font.weight": "bold"})
plt.figure(figsize=(15, 5))
# Plot unmitigated results
plt.subplot(121)
plt.title("Unmitigated")
for i in range(len(all_energies)):
plt.plot(
thetas,
all_energies[i],
"--s",
lw=3,
markersize=10,
markeredgecolor="black",
alpha=0.7,
label=f"$p = {pvals[i]}$",
)
plt.xlabel(r"$\theta$")
plt.ylabel(r"$E(\theta)$")
plt.legend()
plt.subplot(122)
plt.title("Mitigated with ZNE")
# Plot noiseless results
plt.plot(
thetas,
all_energies[0],
"--s",
lw=3,
markersize=10,
markeredgecolor="black",
alpha=0.7,
label="$p = 0.0$",
)
# Plot error-mitigated results
for i in range(len(all_mitigated)):
plt.plot(
thetas,
all_mitigated[i],
"--o",
lw=3,
markersize=10,
markeredgecolor="black",
alpha=0.7,
label=f"$p = {pvals[i + 1]}$",
)
plt.xlabel(r"$\theta$")
plt.legend()
plt.tight_layout()
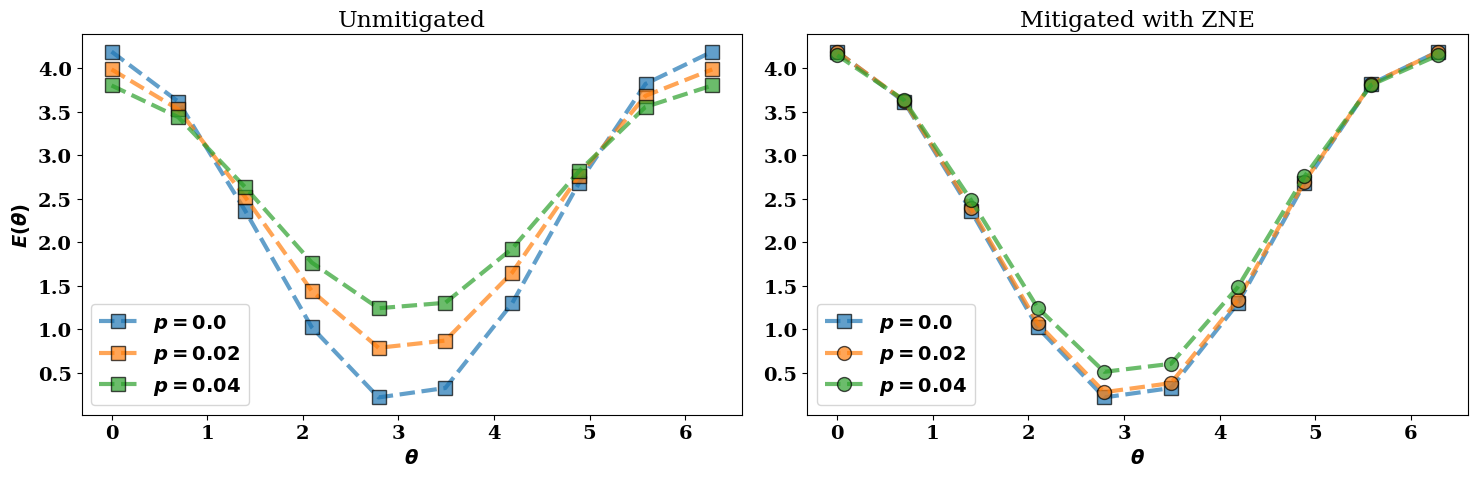
In the figure above, the error-mitigated energy landscapes (colored circles) are closer to the ideal noiseless limit (blue squares) when compared to the unmitigated values (colored squares).
Evaluating the potential energy surface \(V(R)\)#
In the previous section we mitigated the energy landscape \(E(\theta, R)\) for a particular value of \(R\). Here instead we evaluate the potential energy surface, which can be defined as
We first evaluate \(V(R)\) without using error mitigation.
best_thetas = []
best_energies = []
for pval in pvals:
print(f"\nStatus: p = {pval}")
these_thetas = []
these_energies = []
for i in range(len(radii)):
# Objective function to minimize
def obj(theta):
return energy(ansatz(theta[0]), radius_index=i, depo_noise_strength=pval)
res = brute(obj, ranges=[(0, 2 * np.pi)], Ns=10, finish=None, full_output=True)
these_thetas.append(res[0])
these_energies.append(res[1])
best_thetas.append(these_thetas)
best_energies.append(these_energies)
Status: p = 0.0
Status: p = 0.02
Status: p = 0.04
The unmitigated potential energy surfaces are now stored in the best_energies variable and will be visualized later.
Applying zero-noise extrapolation to estimate \(V(R)\)#
We use zero-noise extrapolation to error-mitigate the potential energy surface \(V(R)\).
best_mitigated_thetas = []
best_mitigated_energies = []
for pval in pvals[1:]:
print(f"\nStatus: p = {pval}")
these_thetas = []
these_energies = []
for i in range(len(radii)):
def objective_function(theta):
return zne.execute_with_zne(
ansatz(theta[0]),
partial(energy, radius_index=i, depo_noise_strength=pval),
factory=fac,
scale_noise=scaling_function,
num_to_average=num_to_average,
)
# Minimize energy with respect to "theta"
res = brute(
objective_function,
ranges=[(0, 2 * np.pi)],
Ns=10,
finish=None,
full_output=True,
)
these_thetas.append(res[0])
these_energies.append(res[1])
best_mitigated_thetas.append(these_thetas)
best_mitigated_energies.append(these_energies)
Status: p = 0.02
Status: p = 0.04
In the next figure we visualize the following results:
The ideal noiseless potential energy surface \(V(R)\) corresponding to
best_energies[0];The noisy (unmitigated) potential energy surfaces
best_energies;The corresponding error-mitigated potential energy surfaces
best_mitigated_energies.
plt.figure(figsize=(15, 5))
# Plot unmitigated results
plt.subplot(121)
plt.title("Unmitigated")
for pval, opt_energies in zip(pvals, best_energies):
plt.plot(
radii,
opt_energies,
"--s",
lw=2,
markersize=10,
markeredgecolor="black",
alpha=0.7,
label=f"$p = {pval}$",
)
plt.ylim(
min([np.min(best_energies), np.min(best_mitigated_energies)]) - 0.1,
max([np.max(best_energies), np.max(best_mitigated_energies)]) + 0.1,
)
plt.xlabel("Atomic distance (Angstroms)")
plt.ylabel("Energy (Hartree)")
plt.legend()
# Plot noiseless results
plt.subplot(122)
plt.title("Mitigated with ZNE")
plt.plot(
radii,
best_energies[0],
"-s",
lw=2,
markersize=10,
markeredgecolor="black",
alpha=0.7,
label=f"$p = 0.0$",
zorder=1,
)
# Plot error-mitigated results
i = 0
for pval, opt_energies in zip(pvals[1:], best_mitigated_energies):
i += 1
plt.plot(
radii,
opt_energies,
"-o",
lw=2,
markersize=10,
markeredgecolor="black",
alpha=0.7,
label=f"$p = {pval}$",
zorder=0,
)
plt.ylim(
min([np.min(best_energies), np.min(best_mitigated_energies)]) - 0.1,
max([np.max(best_energies), np.max(best_mitigated_energies)]) + 0.1,
)
plt.xlabel("Atomic distance (Angstroms)")
plt.legend();
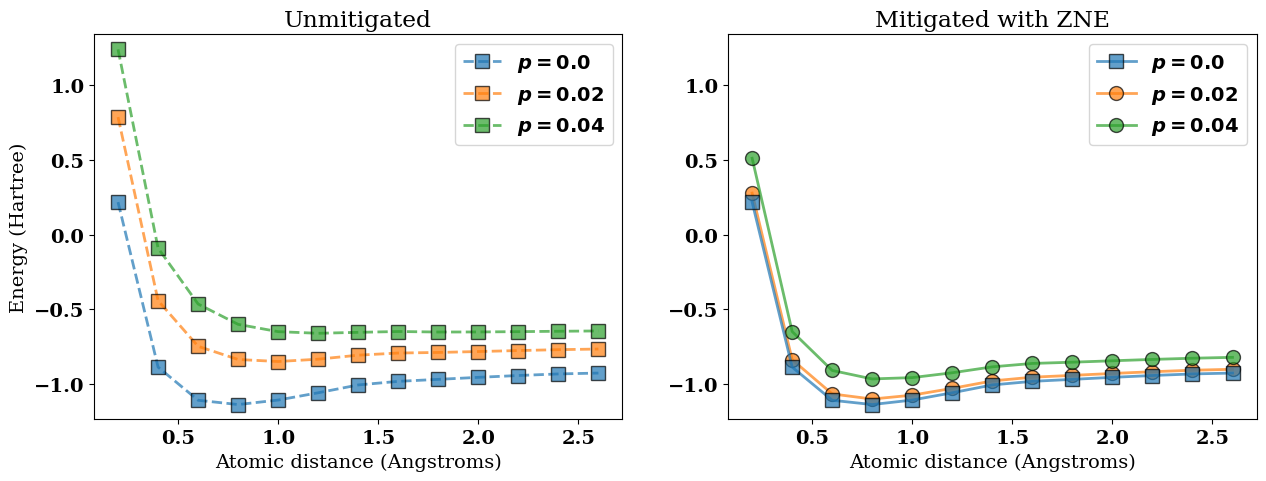
In the figure above, the error-mitigated potential energy surfaces (colored circles) are closer to the ideal noiseless limit (blue squares) when compared to the unmitigated results (colored squares).
